In this Article

Tomato (Solanum Iycopersicum) and pepper (Capsicum annuum) are among the most important vegetables grown by smallholder farmers in the tropics and subtropics. Bacterial wilt (BW), caused by the Ralstonia solanacearum species complex (RSSC), has been reported as the second most important plant pathogenic bacteria worldwide and likely the most destructive. One of the most severe pepper diseases is anthracnose, caused by members of the Colletotrichum species complex (CSC). High temperatures and humidity favor both RSSC and CSC. As extreme weather events become more frequent and severe through climate change, it is anticipated that such diseases will become more common and destructive.
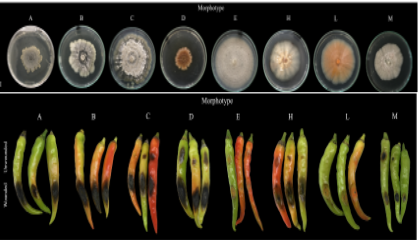
This project aims to develop user-friendly markers to characterize polymorphic loci identified in a panel of highly bacterial wilt resistant and susceptible tomato lines in a population of segregating lines to determine association with the phenotype and inheritance from parents to progeny; scan the pepper genome using advanced bioinformatics techniques to identify regions with high homology to newly identified resistance-associated loci in tomato; and evaluate world vegetable sources of resistance to bacterial wilt and anthracnose.








